Energy evaluation for MFE and partition function calculations. More...
 Include dependency graph for loop_energies.h:
Include dependency graph for loop_energies.h:Go to the source code of this file.
Functions | |
| PRIVATE int | E_IntLoop (int n1, int n2, int type, int type_2, int si1, int sj1, int sp1, int sq1, paramT *P) |
| PRIVATE int | E_Hairpin (int size, int type, int si1, int sj1, const char *string, paramT *P) |
| PRIVATE int | E_Stem (int type, int si1, int sj1, int extLoop, paramT *P) |
| PRIVATE double | exp_E_Stem (int type, int si1, int sj1, int extLoop, pf_paramT *P) |
| PRIVATE double | exp_E_Hairpin (int u, int type, short si1, short sj1, const char *string, pf_paramT *P) |
| PRIVATE double | exp_E_IntLoop (int u1, int u2, int type, int type2, short si1, short sj1, short sp1, short sq1, pf_paramT *P) |
Detailed Description
Energy evaluation for MFE and partition function calculations.
This file contains functions for the calculation of the free energy 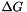 of a hairpin- [ E_Hairpin() ] or interior-loop [ E_IntLoop()] .
of a hairpin- [ E_Hairpin() ] or interior-loop [ E_IntLoop()] .
The unit of the free energy returned is 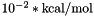
In case of computing the partition function, this file also supplies functions which return the Boltzmann weights 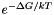 for a hairpin- [ exp_E_Hairpin() ] or interior-loop [ exp_E_IntLoop() ].
for a hairpin- [ exp_E_Hairpin() ] or interior-loop [ exp_E_IntLoop() ].
Function Documentation
| PRIVATE int E_IntLoop | ( | int | n1, |
| int | n2, | ||
| int | type, | ||
| int | type_2, | ||
| int | si1, | ||
| int | sj1, | ||
| int | sp1, | ||
| int | sq1, | ||
| paramT * | P | ||
| ) |
Compute the Energy of an interior-loop
This function computes the free energy 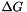 of an interior-loop with the following structure:
of an interior-loop with the following structure:
3' 5'
| |
U - V
a_n b_1
. .
. .
. .
a_1 b_m
X - Y
| |
5' 3'
This general structure depicts an interior-loop that is closed by the base pair (X,Y). The enclosed base pair is (V,U) which leaves the unpaired bases a_1-a_n and b_1-b_n that constitute the loop. In this example, the length of the interior-loop is 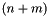 where n or m may be 0 resulting in a bulge-loop or base pair stack. The mismatching nucleotides for the closing pair (X,Y) are:
where n or m may be 0 resulting in a bulge-loop or base pair stack. The mismatching nucleotides for the closing pair (X,Y) are:
5'-mismatch: a_1
3'-mismatch: b_m
and for the enclosed base pair (V,U):
5'-mismatch: b_1
3'-mismatch: a_n
- Note
- Base pairs are always denoted in 5'->3' direction. Thus the enclosed base pair must be 'turned arround' when evaluating the free energy of the interior-loop
- See Also
- scale_parameters()
- paramT
- Note
- This function is threadsafe
- Parameters
-
n1 The size of the 'left'-loop (number of unpaired nucleotides) n2 The size of the 'right'-loop (number of unpaired nucleotides) type The pair type of the base pair closing the interior loop type_2 The pair type of the enclosed base pair si1 The 5'-mismatching nucleotide of the closing pair sj1 The 3'-mismatching nucleotide of the closing pair sp1 The 3'-mismatching nucleotide of the enclosed pair sq1 The 5'-mismatching nucleotide of the enclosed pair P The datastructure containing scaled energy parameters
- Returns
- The Free energy of the Interior-loop in dcal/mol
| PRIVATE int E_Hairpin | ( | int | size, |
| int | type, | ||
| int | si1, | ||
| int | sj1, | ||
| const char * | string, | ||
| paramT * | P | ||
| ) |
Compute the Energy of a hairpin-loop
To evaluate the free energy of a hairpin-loop, several parameters have to be known. A general hairpin-loop has this structure:
a3 a4
a2 a5
a1 a6
X - Y
| |
5' 3'
where X-Y marks the closing pair [e.g. a (G,C) pair]. The length of this loop is 6 as there are six unpaired nucleotides (a1-a6) enclosed by (X,Y). The 5' mismatching nucleotide is a1 while the 3' mismatch is a6. The nucleotide sequence of this loop is "a1.a2.a3.a4.a5.a6"
- Note
- The parameter sequence should contain the sequence of the loop in capital letters of the nucleic acid alphabet if the loop size is below 7. This is useful for unusually stable tri-, tetra- and hexa-loops which are treated differently (based on experimental data) if they are tabulated.
- See Also
- scale_parameters()
- paramT
- Warning
- Not (really) thread safe! A threadsafe implementation will replace this function in a future release!
Energy evaluation may change due to updates in global variable "tetra_loop"
- Parameters
-
size The size of the loop (number of unpaired nucleotides) type The pair type of the base pair closing the hairpin si1 The 5'-mismatching nucleotide sj1 The 3'-mismatching nucleotide string The sequence of the loop P The datastructure containing scaled energy parameters
- Returns
- The Free energy of the Hairpin-loop in dcal/mol
| PRIVATE int E_Stem | ( | int | type, |
| int | si1, | ||
| int | sj1, | ||
| int | extLoop, | ||
| paramT * | P | ||
| ) |
Compute the energy contribution of a stem branching off a loop-region
This function computes the energy contribution of a stem that branches off a loop region. This can be the case in multiloops, when a stem branching off increases the degree of the loop but also immediately interior base pairs of an exterior loop contribute free energy. To switch the bahavior of the function according to the evaluation of a multiloop- or exterior-loop-stem, you pass the flag 'extLoop'. The returned energy contribution consists of a TerminalAU penalty if the pair type is greater than 2, dangling end contributions of mismatching nucleotides adjacent to the stem if only one of the si1, sj1 parameters is greater than 0 and mismatch energies if both mismatching nucleotides are positive values. Thus, to avoid incooperating dangling end or mismatch energies just pass a negative number, e.g. -1 to the mismatch argument.
This is an illustration of how the energy contribution is assembled:
3' 5'
| |
X - Y
5'-si1 sj1-3'
Here, (X,Y) is the base pair that closes the stem that branches off a loop region. The nucleotides si1 and sj1 are the 5'- and 3'- mismatches, respectively. If the base pair type of (X,Y) is greater than 2 (i.e. an A-U or G-U pair, the TerminalAU penalty will be included in the energy contribution returned. If si1 and sj1 are both nonnegative numbers, mismatch energies will also be included. If one of sij or sj1 is a negtive value, only 5' or 3' dangling end contributions are taken into account. To prohibit any of these mismatch contributions to be incoorporated, just pass a negative number to both, si1 and sj1. In case the argument extLoop is 0, the returned energy contribution also includes the internal-loop-penalty of a multiloop stem with closing pair type.
- See Also
- E_MLstem()
- E_ExtLoop()
- Note
- This function is threadsafe
- Parameters
-
type The pair type of the first base pair un the stem si1 The 5'-mismatching nucleotide sj1 The 3'-mismatching nucleotide extLoop A flag that indicates whether the contribution reflects the one of an exterior loop or not P The datastructure containing scaled energy parameters
- Returns
- The Free energy of the branch off the loop in dcal/mol
| PRIVATE double exp_E_Stem | ( | int | type, |
| int | si1, | ||
| int | sj1, | ||
| int | extLoop, | ||
| pf_paramT * | P | ||
| ) |
| PRIVATE double exp_E_Hairpin | ( | int | u, |
| int | type, | ||
| short | si1, | ||
| short | sj1, | ||
| const char * | string, | ||
| pf_paramT * | P | ||
| ) |
Compute Boltzmann weight 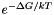 of a hairpin loop
of a hairpin loop
multiply by scale[u+2]
- Warning
- Not (really) thread safe! A threadsafe implementation will replace this function in a future release!
Energy evaluation may change due to updates in global variable "tetra_loop"
- Parameters
-
u The size of the loop (number of unpaired nucleotides) type The pair type of the base pair closing the hairpin si1 The 5'-mismatching nucleotide sj1 The 3'-mismatching nucleotide string The sequence of the loop P The datastructure containing scaled Boltzmann weights of the energy parameters
- Returns
- The Boltzmann weight of the Hairpin-loop
| PRIVATE double exp_E_IntLoop | ( | int | u1, |
| int | u2, | ||
| int | type, | ||
| int | type2, | ||
| short | si1, | ||
| short | sj1, | ||
| short | sp1, | ||
| short | sq1, | ||
| pf_paramT * | P | ||
| ) |
Compute Boltzmann weight 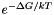 of interior loop
of interior loop
multiply by scale[u1+u2+2] for scaling
- Note
- This function is threadsafe
- Parameters
-
u1 The size of the 'left'-loop (number of unpaired nucleotides) u2 The size of the 'right'-loop (number of unpaired nucleotides) type The pair type of the base pair closing the interior loop type2 The pair type of the enclosed base pair si1 The 5'-mismatching nucleotide of the closing pair sj1 The 3'-mismatching nucleotide of the closing pair sp1 The 3'-mismatching nucleotide of the enclosed pair sq1 The 5'-mismatching nucleotide of the enclosed pair P The datastructure containing scaled Boltzmann weights of the energy parameters
- Returns
- The Boltzmann weight of the Interior-loop