This module contains all functions and variables related to the calculation of global minimum free energy structures for single sequences. More...
 Collaboration diagram for Calculating Minimum Free Energy (MFE) Structures:
Collaboration diagram for Calculating Minimum Free Energy (MFE) Structures:Modules | |
| MFE Structures of two hybridized Sequences | |
| MFE Consensus Structures for Sequence Alignment(s) | |
| Local MFE structure Prediction and Z-scores | |
| Calculating MFE representatives of a Distance Based Partitioning | |
| Compute the minimum free energy (MFE) and secondary structures for a partitioning of the secondary structure space according to the base pair distance to two fixed reference structures basepair distance to two fixed reference structures. | |
Functions | |
| float | fold_par (const char *sequence, char *structure, paramT *parameters, int is_constrained, int is_circular) |
| Compute minimum free energy and an appropriate secondary structure of an RNA sequence. More... | |
| float | fold (const char *sequence, char *structure) |
| Compute minimum free energy and an appropriate secondary structure of an RNA sequence. More... | |
| float | circfold (const char *sequence, char *structure) |
| Compute minimum free energy and an appropriate secondary structure of a circular RNA sequence. More... | |
| void | free_arrays (void) |
| Free arrays for mfe folding. | |
| void | update_fold_params (void) |
| Recalculate energy parameters. | |
Detailed Description
This module contains all functions and variables related to the calculation of global minimum free energy structures for single sequences.
This section covers all functions and variables related to the calculation of minimum free energy (MFE) structures.
The library provides a fast dynamic programming minimum free energy folding algorithm as described by Zuker & Stiegler (1981).
The library provides a fast dynamic programming minimum free energy folding algorithm as described in[14]. All relevant parts that directly implement the "Zuker & Stiegler" algorithm for single sequences are described in this section.
Folding of circular RNA sequences is handled as a post-processing step of the forward recursions. See[4] for further details.
Nevertheless, the RNAlib also provides interfaces for the prediction of consensus MFE structures of sequence alignments, MFE structure for two hybridized sequences, local optimal structures and many more. For those more specialized variants of MFE folding routines, please consult the appropriate subsections (Modules) as listed above.
Function Documentation
| float fold_par | ( | const char * | sequence, |
| char * | structure, | ||
| paramT * | parameters, | ||
| int | is_constrained, | ||
| int | is_circular | ||
| ) |
Compute minimum free energy and an appropriate secondary structure of an RNA sequence.
The first parameter given, the RNA sequence, must be uppercase and should only contain an alphabet 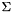 that is understood by the RNAlib
that is understood by the RNAlib
(e.g. 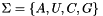 )
)
The second parameter, structure, must always point to an allocated block of memory with a size of at least 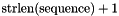
If the third parameter is NULL, global model detail settings are assumed for the folding recursions. Otherwise, the provided parameters are used.
The fourth parameter indicates whether a secondary structure constraint in enhanced dot-bracket notation is passed through the structure parameter or not. If so, the characters " | x < > " are recognized to mark bases that are paired, unpaired, paired upstream, or downstream, respectively. Matching brackets " ( ) " denote base pairs, dots "." are used for unconstrained bases.
To indicate that the RNA sequence is circular and thus has to be post-processed, set the last parameter to non-zero
After a successful call of fold_par(), a backtracked secondary structure (in dot-bracket notation) that exhibits the minimum of free energy will be written to the memory structure is pointing to. The function returns the minimum of free energy for any fold of the sequence given.
- Note
- OpenMP: Passing NULL to the 'parameters' argument involves access to several global model detail variables and thus is not to be considered threadsafe
- See Also
- fold(), circfold(), model_detailsT, set_energy_model(), get_scaled_parameters()
- Parameters
-
sequence RNA sequence structure A pointer to the character array where the secondary structure in dot-bracket notation will be written to parameters A data structure containing the prescaled energy contributions and the model details. (NULL may be passed, see OpenMP notes above) is_constrained Switch to indicate that a structure contraint is passed via the structure argument (0==off) is_circular Switch to (de-)activate postprocessing steps in case RNA sequence is circular (0==off)
- Returns
- the minimum free energy (MFE) in kcal/mol
| float fold | ( | const char * | sequence, |
| char * | structure | ||
| ) |
Compute minimum free energy and an appropriate secondary structure of an RNA sequence.
This function essentially does the same thing as fold_par(). However, it takes its model details, i.e. temperature, dangles, tetra_loop, noGU, no_closingGU, fold_constrained, noLonelyPairs from the current global settings within the library
Use fold_par() for a completely threadsafe variant
- See Also
- fold_par(), circfold()
- Parameters
-
sequence RNA sequence structure A pointer to the character array where the secondary structure in dot-bracket notation will be written to
- Returns
- the minimum free energy (MFE) in kcal/mol
| float circfold | ( | const char * | sequence, |
| char * | structure | ||
| ) |
Compute minimum free energy and an appropriate secondary structure of a circular RNA sequence.
This function essentially does the same thing as fold_par(). However, it takes its model details, i.e. temperature, dangles, tetra_loop, noGU, no_closingGU, fold_constrained, noLonelyPairs from the current global settings within the library
Use fold_par() for a completely threadsafe variant
- See Also
- fold_par(), circfold()
- Parameters
-
sequence RNA sequence structure A pointer to the character array where the secondary structure in dot-bracket notation will be written to
- Returns
- the minimum free energy (MFE) in kcal/mol

