rRNAFilter V1.1 Manual
1. Prerequisites
rRNAFilter can be run on Linux and Windows with Java and KAnalyze installed:** Java
You need to install Java 7 (64-bit is highly recommended). To check the version of the Java installed, please input the following command in the terminal:
java -versionThe latest version can be downloaded here.
** KAnalyze
We use KAnalyze to do the k-mer counting.
This tool was included in the software package.
Reference: Audano, Peter, and Fredrik Vannberg. "KAnalyze: a fast versatile pipelined K-mer toolkit." Bioinformatics 30.14 (2014): 2070-2072.
The KAnalyze commands are in the same folder with rRNAFilter command.
2. Input Parameters
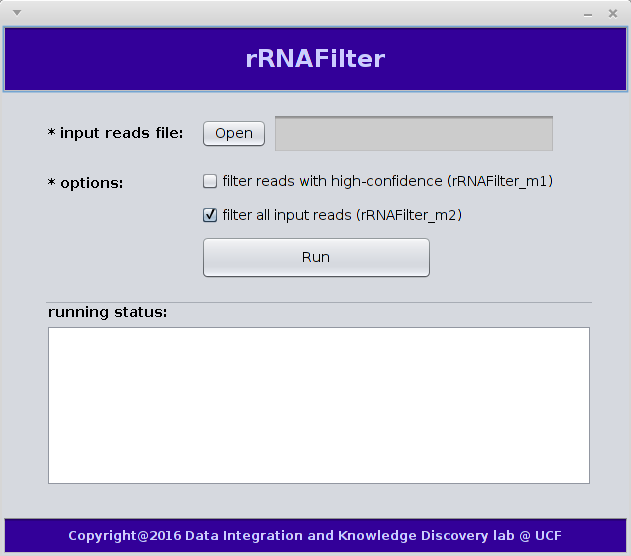
We just need to input reads in fasta format from metatranscirptomes to run rRNAFilter:There are two modes to run rRNAFilter:
- rRNAFilter_m1:filter rRNA reads confidently, and leave a small number of reads unfiltered.
- rRNAFilter_m2:filter all rRNA reads (default option)
3. Software Usage
We have both command line and GUI versions of rRNAFilter on Linux and Windows.(1) In terminal, use the following command to run rRNAFilter:
java -jar -XmxN rRNAFilter.jar -i reads_file -r 0
- -XmxN(for example:-Xmx7g): Specify the maximum size of the memory allocation pool for a Java Virtual Machine (JVM)
- -i (required) reads_file: file in FASTA format
- -r (optional) 0: filter all reads; 1: filter reads confidently, by default r=0
- r=0: filter rRNA reads from all reads.
- r=1: filter rRNA reads initially, without the step to assign the small number of unfiltered reads to the trained Markov chains. So there are still a small number of reads unfiltered.
4. rRNAFilter Results
rRNAFilter will output two files, one file contains predicted rRNA reads and the other file contains predicted non-rRNA reads, the output file names are listed below:- readsFile_rRNA
- readsFile_non-rRNA
- readsFile_non_filtered (if r=1, this is the group of unfiltered reads)